Structured illumination#
abTEM supports various kinds of structured illumination for use with focused or parallel probes.
The code examples below were contributed by Stephanie Ribet based on work published in [RZV+23] and [ZMullerB+20].
Let’s first define a standard focused probe.
energy = 300e3
semiangle_cutoff = 5
extent = [128,128]
sampling = [0.2,0.2]
probe = abtem.Probe(
energy = energy,
extent = extent,
sampling = sampling,
semiangle_cutoff = semiangle_cutoff
);
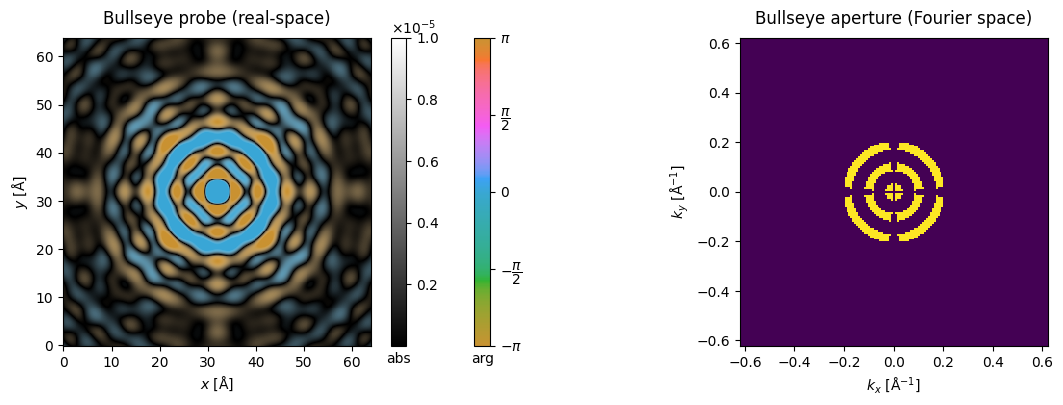
Bullseye aperture#
So-called bullseye apertures can be defined with a dedicated transfer class in abTEM, with a given number and width (in mrad) of “spokes” (radial connecting elements) and “rings” (azimuthal elements). In practice this is used as an aperture in Probe to define a bullseye probe.
bullseye = abtem.transfer.Bullseye(
num_spokes = 4,
num_rings = 3,
spoke_width = 10,
ring_width = 1,
semiangle_cutoff = semiangle_cutoff,
energy = energy,
extent = extent,
sampling = sampling
)
probe_bullseye = abtem.Probe(
energy = energy,
extent = extent,
sampling = sampling,
semiangle_cutoff = semiangle_cutoff,
aperture = bullseye
)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14,4))
probe_bullseye.build().to_images().crop((64,64),(32,32)).show(ax=ax1, vmax = 1e-5, cbar=True, title="Bullseye probe (real-space)")
probe_bullseye.build().diffraction_patterns(max_angle=12).show(ax=ax2, title="Bullseye aperture (Fourier space)");
4D-STEM with bullseye probe#
Let’s run an example simulation with a Ni slab.
Note that the real-space extent of the potential determines the reciprocal-space sampling of the diffraction patterns, hence the large number of repetitions!
Ni_atoms = read("data/Ni.cif")
Ni_atoms = surface(Ni_atoms, (1, 1, 0), 4, periodic=True)
repetitions = (10,14,12)
Ni_atoms *=repetitions
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(14,4))
abtem.show_atoms(Ni_atoms, ax=ax1, title='Top view')
abtem.show_atoms(Ni_atoms, ax=ax2, plane='yz', title='Side view')
abtem.show_atoms(Ni_atoms, ax=ax3, plane='xz', title='Side view');
Adding frozen phonons to the potential; if we did not use them we could use CrystalPotential instead of repeating the model above.
frozen_phonons = abtem.FrozenPhonons(Ni_atoms, 12, 0.1)
potential = abtem.Potential(
frozen_phonons,
sampling=0.2
)
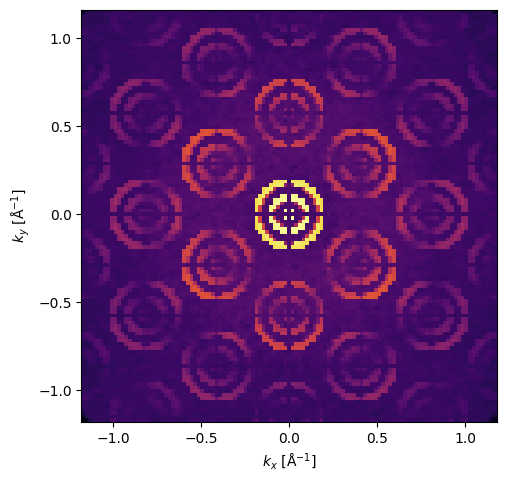
Scanning a grid over one unit cell of the potential with a full 2D pixelated detector.
scan_end = (potential.extent[0] / repetitions[0], potential.extent[1] / repetitions[1]) # Scan over one unit cell.
gridscan = abtem.GridScan(start=[0, 0], end=scan_end, sampling=1.0)
detector = abtem.PixelatedDetector(max_angle='valid', resample='uniform')
measurement = probe_bullseye.scan(
potential=potential,
scan=gridscan,
detectors=detector,
).compute()
A single diffraction pattern selected from the scan shows the effect of the aperture.
measurement[0, 0].show(cmap='inferno', power=0.2);
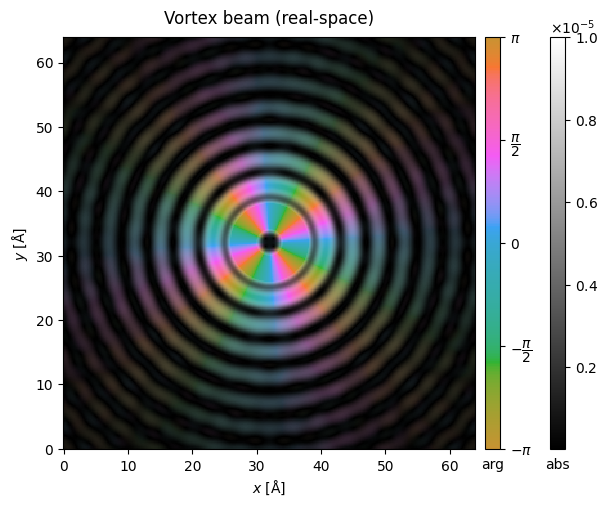
Vortex beams#
We can also define vortex beams with a given quantum number, resulting in variation of the complex phase around the azimuthal direction of the probe.
vortex = abtem.transfer.Vortex(
quantum_number = 4,
semiangle_cutoff = semiangle_cutoff,
energy = energy,
extent = extent,
sampling = sampling
)
probe_vortex = abtem.Probe(
energy = energy,
extent = extent,
sampling = sampling,
aperture = vortex,
semiangle_cutoff = semiangle_cutoff
)
probe_vortex.build().to_images().crop((64,64),(32,32)).show(vmax = 1e-5, cbar=True, title="Vortex beam (real-space)");
TEM phase plates#
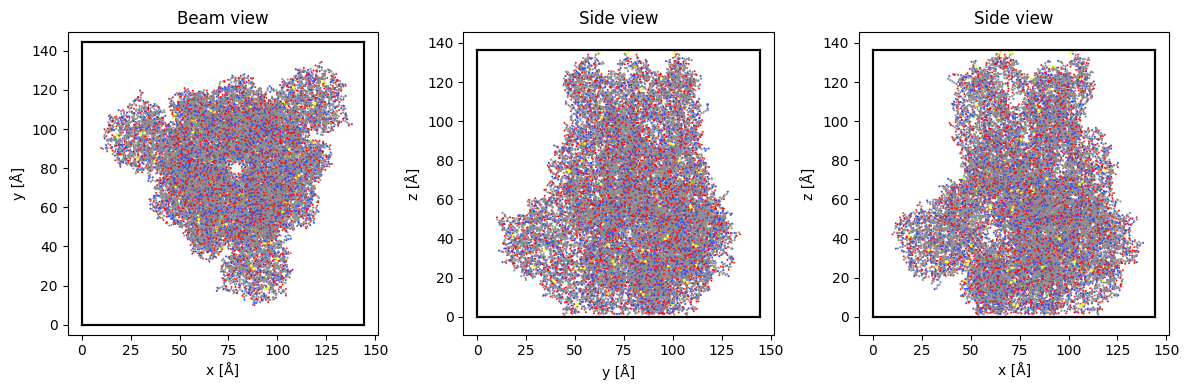
Sample: Covid spike protein
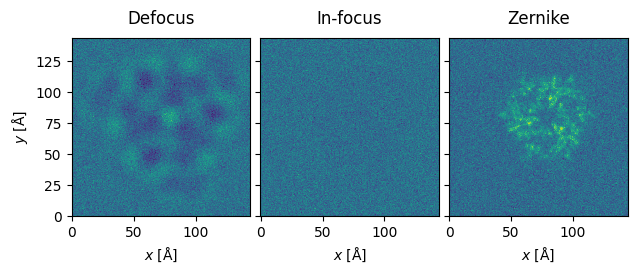
Finally, let’s illustrate the use of a Zernike phase plate on an HRTEM phase-contrast image of the Covid spike protein.
#Load protein structure.
atoms = read('data/3jcl.xyz')
atoms.positions[:,0] -= atoms.positions[:,0].min()
atoms.positions[:,1] -= atoms.positions[:,1].min()
atoms.positions[:,2] -= atoms.positions[:,2].min()
atoms.cell[0][0] = atoms.positions[:,0].max()
atoms.cell[1][1] = atoms.positions[:,1].max()
atoms.cell[2][2] = atoms.positions[:,2].max()
atoms.center(vacuum = 10, axis = (0,1))
atoms.cell[0][0] = atoms.cell[1][1]
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(12, 4))
abtem.show_atoms(atoms, ax=ax1, title="Beam view", linewidth=0)
abtem.show_atoms(atoms, ax=ax2, plane="yz", title="Side view", linewidth=0)
abtem.show_atoms(atoms, ax=ax3, plane="xz", title="Side view", linewidth=0)
fig.tight_layout();
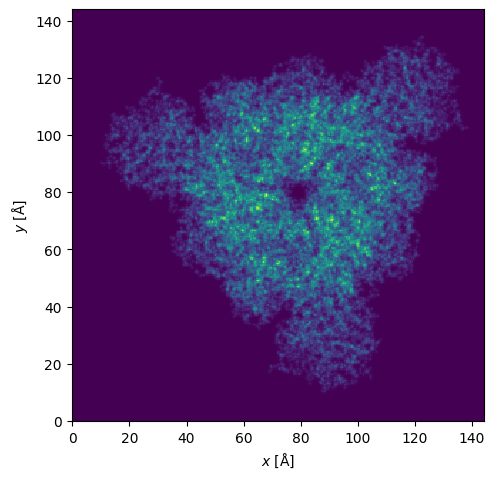
#Make and show the potential.
potential = abtem.Potential(
atoms,
sampling = 0.5,
)
potential.project().show();
#Calculate exit waves.
wave = abtem.waves.PlaneWave(energy=300e3)
exit_waves = wave.multislice(potential).compute()
Let’s then define a Zernike phase plate (which is a special instance of a more generic RadialPhasePlate class): a central hole surrounded by another annular area up to the semiangle cutoff with the given phase shift of \(\pi\).
zernike = abtem.transfer.Zernike(
center_hole_cutoff = 1/10,
phase_shift = np.pi,
semiangle_cutoff = semiangle_cutoff,
energy = energy,
)
#CTFs for in-focus and defocus.
ctf_defocused = abtem.CTF(
aberration_coefficients={'C10': -10000},
semiangle_cutoff = 1,
energy=300e3)
ctf_infocus = abtem.CTF(
aberration_coefficients={'C10': 0},
semiangle_cutoff = 1,
energy=300e3)
dose = 1000
#Calculate in-focus, defocus, and Zernike images.
image_defocus = exit_waves.apply_ctf(ctf_defocused)
image_infocus = exit_waves.apply_ctf(ctf_infocus)
image_zernike = exit_waves.apply_ctf(zernike)
intensity_defocus = image_defocus.intensity()
intensity_infocus = image_infocus.intensity()
intensity_zernike = image_zernike.intensity()
noisy_defocus = intensity_defocus.poisson_noise(dose)
noisy_infocus = intensity_infocus.poisson_noise(dose)
noisy_zernike = intensity_zernike.poisson_noise(dose)
#Zernike provides best transfer of information
noisy_stack = abtem.stack([noisy_defocus, noisy_infocus, noisy_zernike], ["Defocus","In-focus","Zernike"])
noisy_stack.show(explode=True, common_color_scale=True);